Our work on “Wide kernels and their DCT compression in convolutional networks for nuclei segmentation“, V. Andrearczyk, V. Oreiller and A. Depeursinge, has been published in Informatics in Medicine Unlocked.
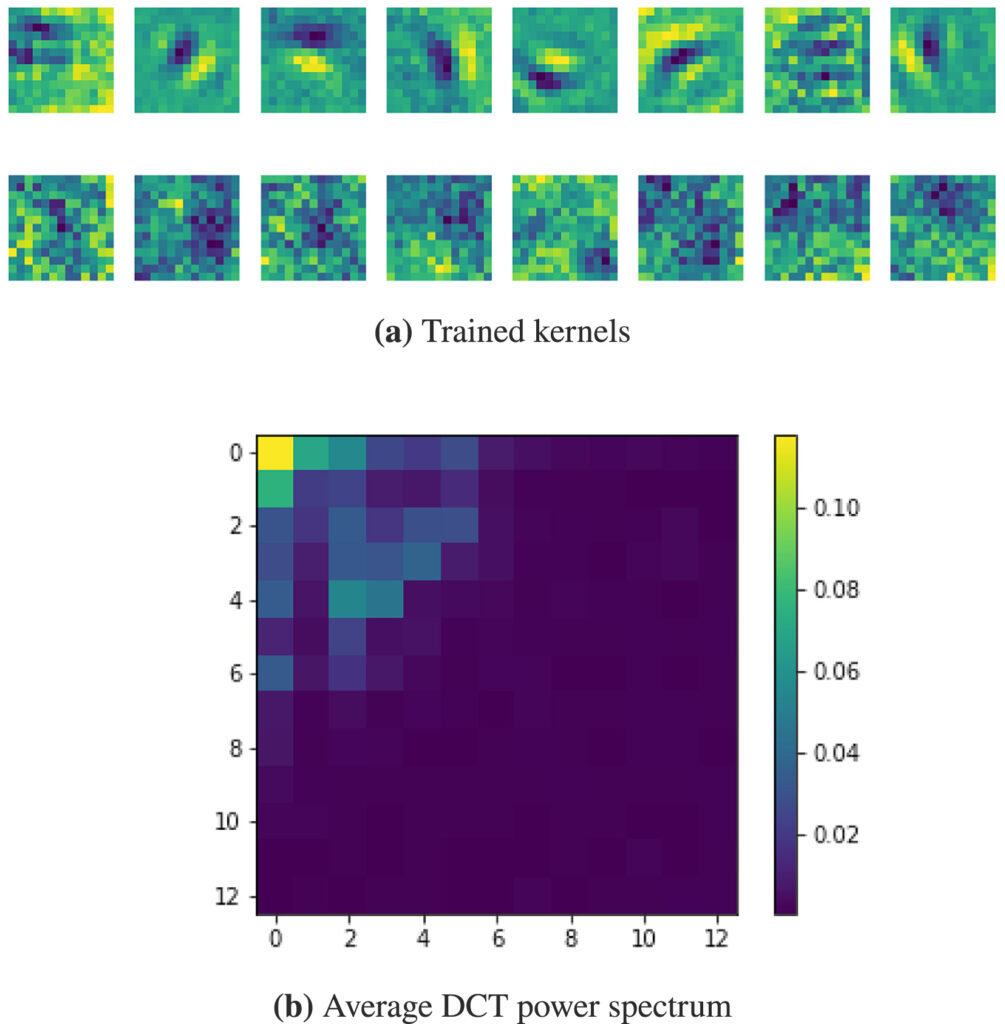
In Convolutional Neural Networks (CNNs), the field of view is traditionally set to very small values (e.g. 3 × 3 pixels) for individual kernels and grown throughout the network by cascading layers. Automatically learning or adapting the best spatial support of the kernels can be done by using large kernels. Obtaining an optimal receptive field with few layers is very relevant in applications with a limited amount of annotated training data, e.g. in medical imaging.
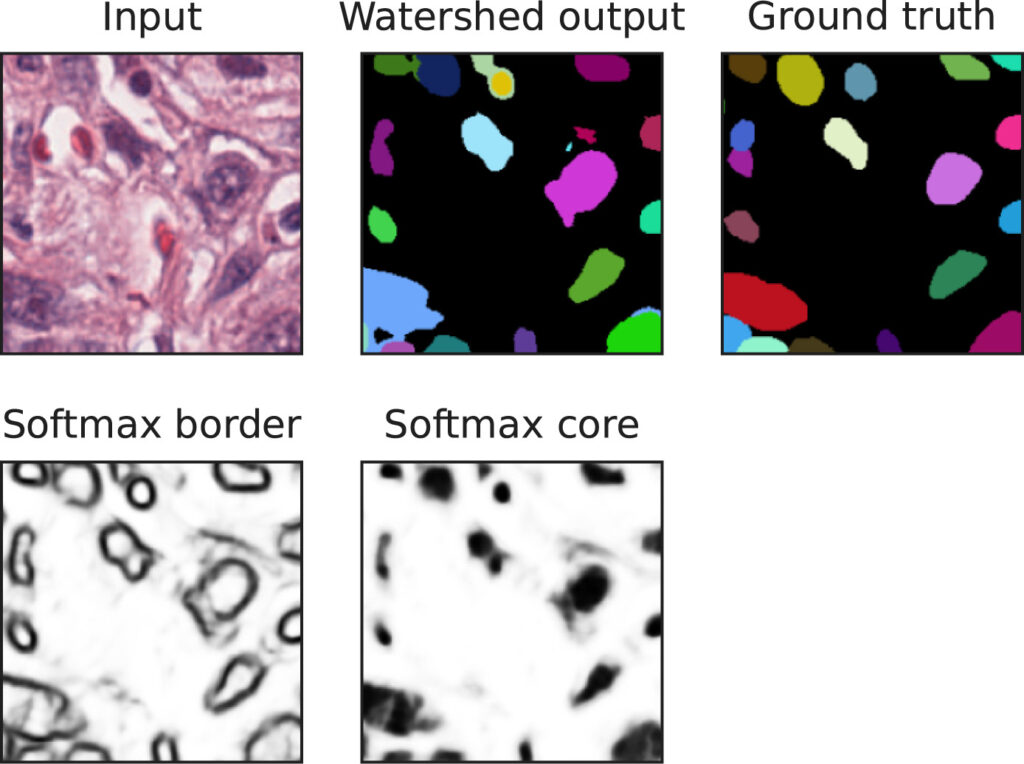
We show that CNNs (2D U-Nets) with large kernels outperform similar models with standard small kernels on the task of nuclei segmentation in histopathology images. We observe that the large kernels mostly capture low-frequency information, which motivates the need for large kernels and their efficient compression via the Discrete Cosine Transform (DCT). Following this idea, we develop a U-Net model with wide and compressed DCT kernels that leads to similar performance and trends to the standard U-Net, with reduced complexity.