Our article entitled “Unleashing the potential of digital pathology data by training computer-aided diagnosis models without human annotations“, by Niccolò Marini et al. has been published in npj Digital Medicine from Nature Portfolio.
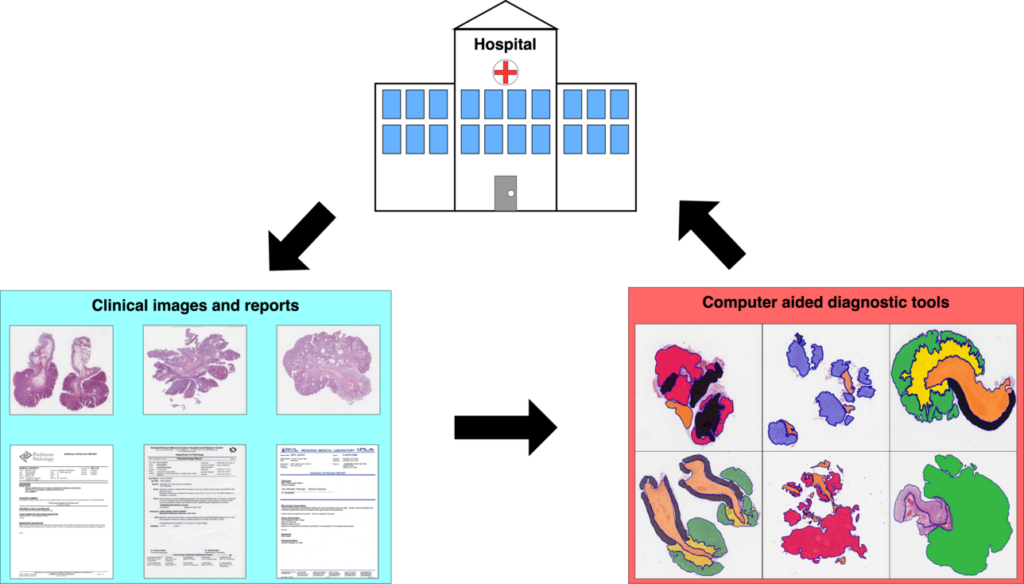
In this work, we present an approach that removes the need for medical experts when building assisting tools for clinical decision support, paving the way for the exploitation of exa-scale sources of medical data collected worldwide from hospital workflows.
The combination of large amounts of healthcare data with new artificial intelligence technologies are leading to the creation of new systems to assist medical experts during the diagnosis.
However, the potential of the combination is not fully exploited because of a bottleneck represented by the need of medical experts to analyze and annotate healthcare data.
This study presents an approach aiming to remove the need of human experts to annotate data: the reports corresponding to healthcare data (that can be CT scans, MRI, whole slide images) are automatically analyzed in order to create automatic annotations that can be used to train deep learning models.
The approach involves two components: a natural language processing algorithm, called SKET (Semantic Knowledge Extractor Tool) that analyzes the reports and extracts meaningful concepts, and a computer vision algorithm, a convolutional neural network trained to classify medical images using the concepts extracted from SKET as weak labels.
The code is available in our GitHub repository for the computer vision algorithm and the natural language processing algorithm.