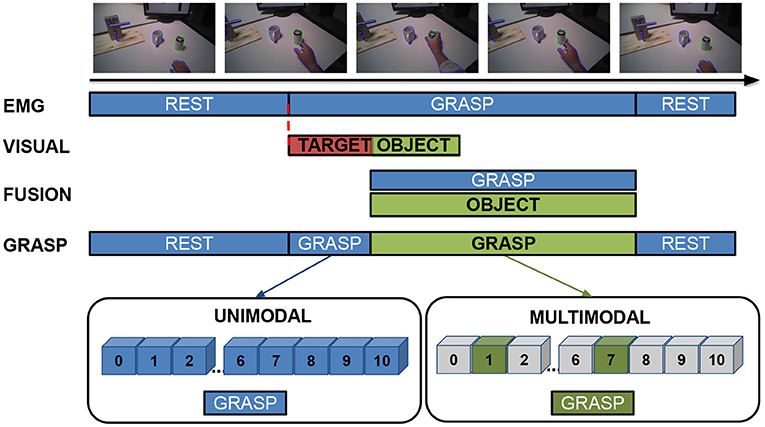
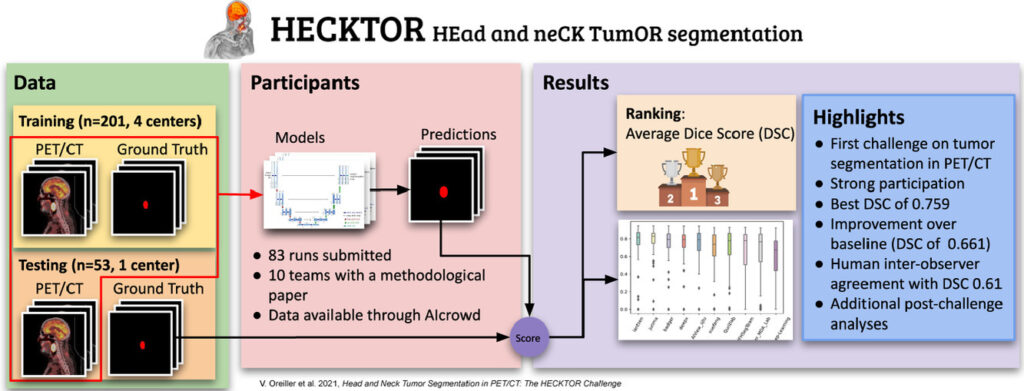
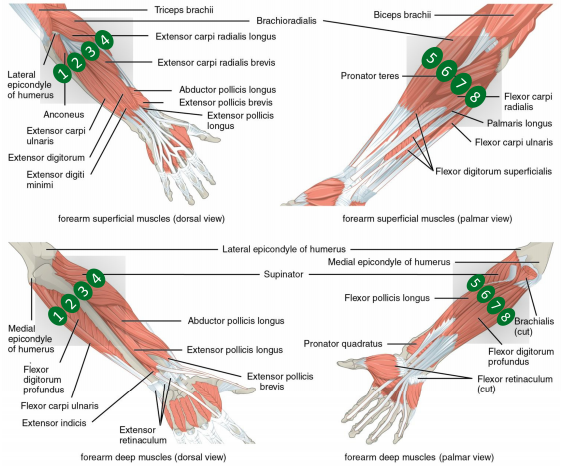
In 2021, the Swiss innovation agency Innosuisse has launched the funding scheme Flagship aimed at stimulating systemic innovation and transdisciplinary collaboration to solve future challenges relevant for the Swiss economy and society. Among the 15 supported projects out of 84 submitted, Innosuisse has financed with 11.2 MCHF over 5 years the project SwissNeuroRehab.
As part of a large Swiss consortium working on this project, our group at the University of Applied Science of Western Switzerland (HES-SO) will lead the “Data” sub project.
SwissNeuroRehab aims at developing a novel model of neurorehabilitation along the continuum of care, from the hospital to home. In the first phase, the project will focus on stroke, traumatic brain injury and spinal cord injury. Through the partnership between university hospitals, research centers, neurorehabilitation clinics, therapists and the industry, SwissNeuroRehab will combine the best available approaches for neuro-rehabilitation with new digital and techonological methods to create innovative and efficient therapeutic programs tailored to the individual needs of patients and their
families.
More details available here (in french).