Master’s student
Darya Ardan
Open PhD position – Medical Signal and image analysis
We are looking for a talented PhD student in the MedGIFT research group at HES-SO in Sierre, Switzerland.
The position involves processing and analyzing neuroimaging data from various modalities and integrating them with biosensor data to aid in early stroke diagnosis, treatment planning, and prognosis.
The PhD will be part of a four-year research project in collaboration with the Hospital of Valais in Sion and the University of Geneva.
Please find the official position announcement here.
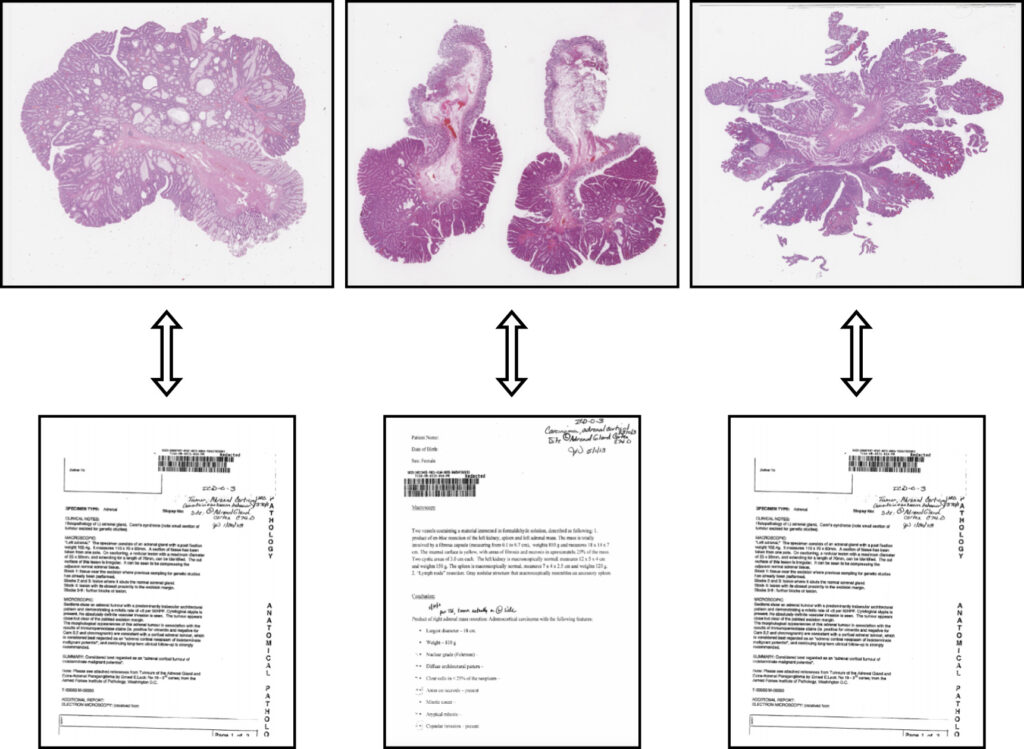
New Multimodal Architecture for Computational Pathology published in Medical Image Analysis
Our article “Multimodal representations of biomedical knowledge from limited training whole slide images and reports using deep learning“, Niccolò Marini et al., has been published in Medical Image Analysis.
In this work, we developed a novel multimodal learning architecture that integrates images and reports, offering a robust new approach to histopathology data representation. This model can be used as a powerful backbone to solve various computational pathology tasks.
The multimodal data representation outperforms unimodal representations in terms of whole slide image (WSI) classification, making it possible to exploit smaller datasets to train effective networks.
The combination of unsupervised learning methods allows it to learn similarities and dissimilarities between images and reports. The model effectively represents medical ontologies through visual knowledge, creating meaningful connections between the visual information in images and the textual details in reports. This integration opens up new possibilities for enhancing the accuracy and efficiency of computational pathology, which can contribute to better patient outcomes and advancing the field of medical research.
Two papers accepted at MIE2024
Two of our papers were accepted for presentation at MI2024, 34th Medical Informatics Europe Conference, which will take place from August 25-29 in Athens, Greece.
“An Overview of Public Retinal Optical Coherence Tomography Datasets: Access, Annotations, and Beyond”, Anastasiia Rozhyna et al.
In ophthalmology, Optical Coherence Tomography (OCT) is a daily diagnostic and therapeutic tool for various diseases. Publicly available datasets are crucial for research but suffer from inconsistent accessibility, data formats, annotations, and metadata. This article analyzes different OCT datasets, focusing on properties, disease representation, and accessibility, aiming to catalog all public OCT datasets. The goal is to improve data accessibility, transparency, and provide new perspectives on OCT imaging resources.
“PICO to PICOS: Weak Supervision to Extend Datasets with New Labels”, Anjani Dhrangadhariya et al.
Using a weakly supervised approach, this work achieved an impressive F1 score of 85.02% in extracting clinical entities without labelled data. Leveraging data programming and resources like UMLS and NCBOBioportal, the workflow successfully extracts “Study type and design” from clinical trial abstracts in the EBM PICO dataset.

Tutorial on radiomics at EUVIP 2024 (September 8)
Adrien Depeursinge, Mario Jreige (CHUV) and Vincent Andrearczyk will give a tutorial on Radiomics: success stories, negative results, challenges ahead and hands-on sessions, at the European Workshop on Visual Information Processing (EUVIP) 2024, Sunday, 8th September in Geneva University Hospital.
Registration is now open here.
The first part of this tutorial will review the most notable achievements to date, including FDA and CE-approved products addressing image-based personalized medicine, clinical adoption, as well as notable negative results and areas where initial promises have not been fulfilled. We will then summarize best practices, study quality assessment, and identify paradigms most appropriate for building successful radiomics models. Additionally, we will share our experience in the domain, covering the organization of radiomics challenges (HECKTOR), the Image Biomarker Standardisation Initiative (IBSI), and the design and use of anthropomorphic phantoms to assess the reproducibility and harmonization of radiomics.
In the second part, we will organize hands-on sessions where participants will have the opportunity to develop, validate, and interpret radiomics models.
We will conclude the tutorial with a participatory discussion on the hands-on experience, followed by general concluding remarks.
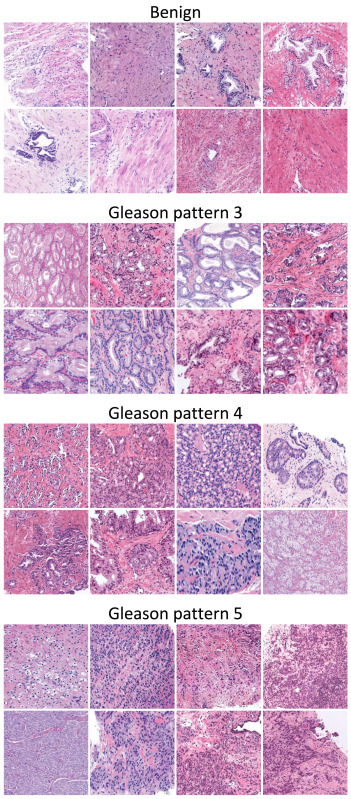
Article on prostate cancer grading published in Medical Image Analysis
Our article entitled “A systematic comparison of deep learning methods for Gleason grading and scoring“, J. P. Dominguez-Morales et al., has been published in Medical Image analysis.
Prostate cancer ranks second among men’s cancers globally, following lung cancer. Diagnosis relies on the Gleason score, assessing cell abnormality in glandular tissue through various patterns. Computational pathology advancements offer diverse datasets and algorithms for Gleason grading, yet consensus lacks on optimal methods matching data and labels.
This paper aims to guide researchers in selecting the most suitable practices based on the task and available labels. It systematically compares, on nine datasets, state-of-the-art training approaches for deep neural networks covering fully-supervised, weakly-supervised, semi-supervised, and various MIL methods. The results indicate that fully supervised learning is best suited for Gleason grading tasks, while Clustering-constrained Attention Multiple Instance Learning (CLAM) performs best for Gleason scoring.
AI for the prediction of major adverse cardiovascular events
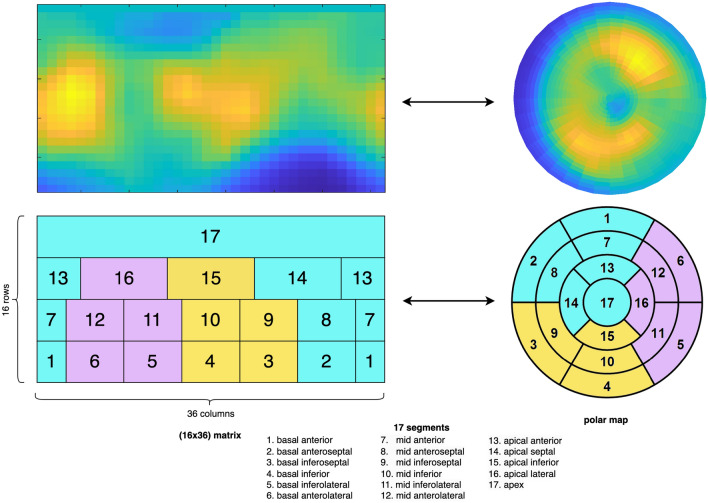
Our work on AI for assessing myocardial perfusion in [82Rb] PET images for the prediction of major adverse cardiovascular events (MACE) has been published in Nature Scientific Reports.
In this article, entitled “Comparing various AI approaches to traditional quantitative assessment of the myocardial perfusion in [82Rb] PET for MACE prediction“, Sacha Bors et al., we show that AI models can allow better personalized prognosis assessment for MACE.
Assessing the individual risk of Major Adverse Cardiac Events (MACE) is of major importance as cardiovascular diseases remain the leading cause of death worldwide. Quantitative Myocardial Perfusion Imaging (MPI) parameters such as stress Myocardial Blood Flow (sMBF) or Myocardial Flow Reserve (MFR) constitute the gold standard for prognosis assessment. We propose a systematic investigation of the value of Artificial Intelligence (AI) to leverage [82Rb] PET MPI for MACE prediction. We establish a general pipeline for AI model validation to assess and compare the performance of global (i.e. average of the entire MPI signal), regional (17 segments), radiomics and Convolutional Neural Network (CNN) models leveraging various MPI signals on a dataset of 234 patients. Results showed that all regional AI models significantly outperformed the global model (p<0.001), where the best AUC of 73.9% was obtained with a CNN model. A regional AI model based on MBF averages from 17 segments fed to a Logistic Regression (LR) constituted an excellent trade-off between model simplicity and performance, achieving an AUC of 73.4%. A radiomics model based on intensity features revealed that the global average was the least important feature when compared to other aggregations of the MPI signal over the myocardium.
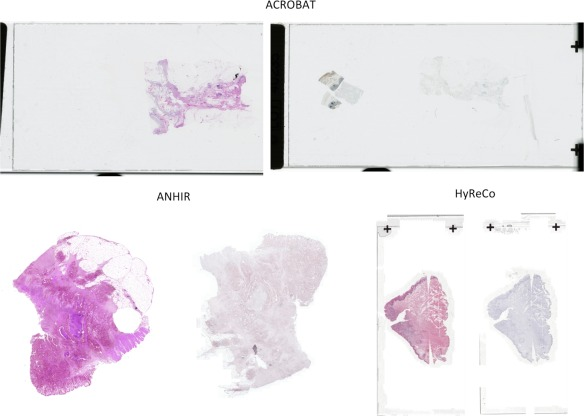
Article published on Whole Slide Images registration
Our work on Whole Slide Images (WSIs) registration was published in Computer Methods and Programs in Biomedicine:
“RegWSI: Whole slide image registration using combined deep feature- and intensity-based methods: Winner of the ACROBAT 2023 challenge“, by M. Wodzinski, N. Marini, M. Atzori, and H. Müller
In this article, we introduce a new method dedicated to automatic registration of WSIs. The proposed method has superior generalizability and does not require any re-training or fine-tuning to particular dataset. The quantitative results are very accurate, and the algorithm is the best registration method on the ACROBAT and HyReCo datasets.
The source code is released and included in the DeeperHistReg framework, allowing end-users to use it in their research.
Open PhD position – Machine learning and multimedia knowledge extraction from biomedical data
We are looking for a talented PhD student in the MedGIFT research group at HES-SO in Sierre, Switzerland.
The position involves working on machine learning and multimedia knowledge extraction from biomedical data with special emphasis on decision support, deep neural networks, explainable AI and user tests.
The PhD will be part of a research project funded by the Swiss national science foundation (SNSF) for four years.
Please find the official position announcement here.
Presentation of V. Andrearczyk on AI for medical imaging
Vincent Andrearczyk presented his research on medical imaging at the AI-Cafe, an online forum to gain insights into the European AI scene. He emphasized the importance of three essential ingredients: Generalizability, interpretability and interaction with clinicians. The talk is available online:
Medical imaging is an essential step in patient care, from diagnostic and treatment planning to follow-up, allowing doctors to assess organs, tissue and blood vessels non-invasively. AI capabilities to analyze medical images are extremely promising for assisting clinicians in their daily routines.
This presentation introduces some of the essential ingredients for developing reliable medical imaging AI models with a focus on generalizability, interpretability and interaction with clinicians.
Generalizability refers to the capacity of the models to adapt to new, previously unseen data, for instance, images coming from a new machine or hospital. Interpretability refers to the translation of the working principles and outcomes of the models in human-understandable terms. Finally, the involvement of clinicians, in all phases of a model development and evaluation is crucial to ensure the utility, usability and alignment of the solutions.
This talk covered all these topics and their integration in various tasks to foster patient care. I will give concrete examples including brain lesion management based on MRI analysis, and head and neck tumor segmentation and outcome prediction from PET/CT images.
Review article on XAI in medical imaging
Our review article “A Scoping Review of Interpretability and Explainability concerning Artificial Intelligence Methods in Medical Imaging“, M. Champendal, H. Müller, J. O. Prior and C. Sá dos Reis, has been published in the European Journal of Radiology.
Our study shows the increase of XAI publications, primarily focusing on applying MRI, CT, and radiography for classifying and predicting lung and brain pathologies. Visual and numerical output formats are predominantly used. We also show that terminology standardisation remains a challenge, as terms like “explainable” and “interpretable” are sometimes used indistinctively. More details and interesting results are available in the full paper.

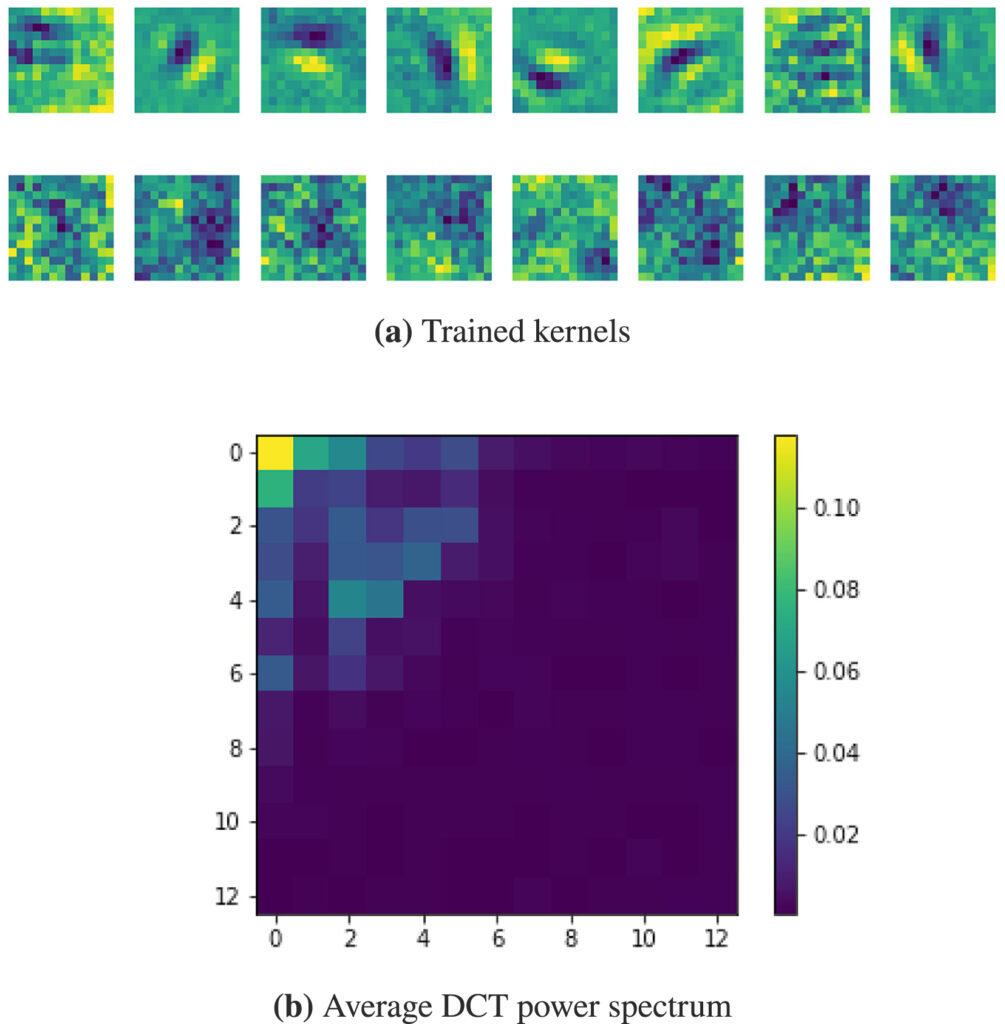
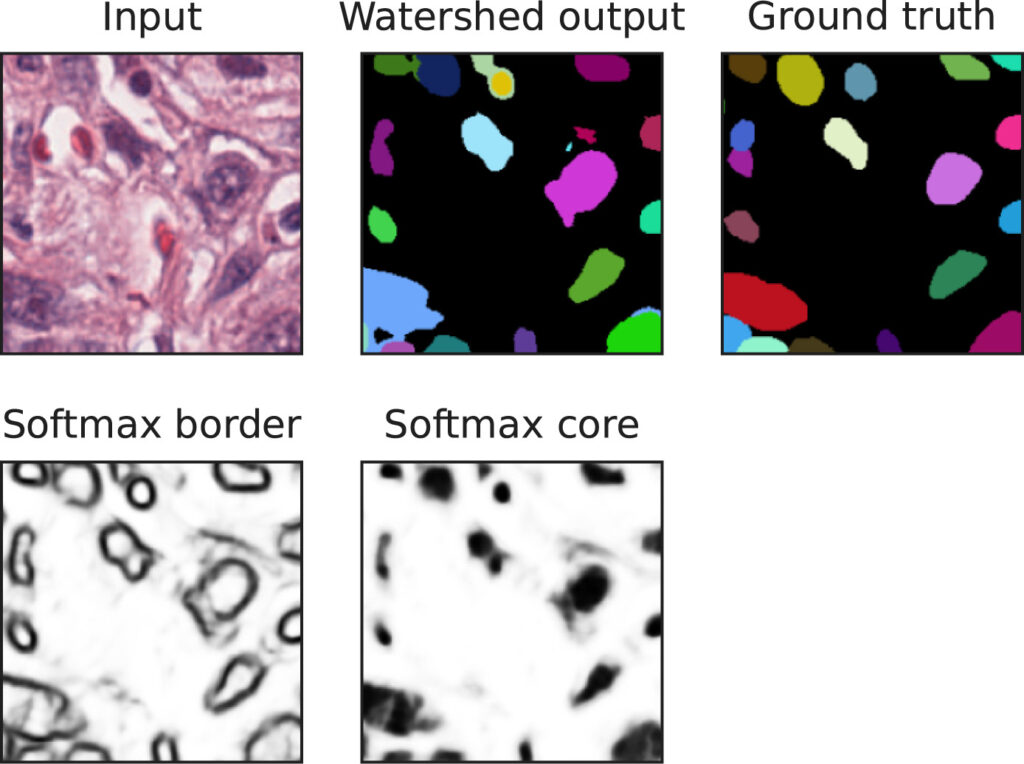
Article on wide kernels and DCT in CNNs published in Informatics in Medicine Unlocked
Our work on “Wide kernels and their DCT compression in convolutional networks for nuclei segmentation“, V. Andrearczyk, V. Oreiller and A. Depeursinge, has been published in Informatics in Medicine Unlocked.
In Convolutional Neural Networks (CNNs), the field of view is traditionally set to very small values (e.g. 3 × 3 pixels) for individual kernels and grown throughout the network by cascading layers. Automatically learning or adapting the best spatial support of the kernels can be done by using large kernels. Obtaining an optimal receptive field with few layers is very relevant in applications with a limited amount of annotated training data, e.g. in medical imaging.
We show that CNNs (2D U-Nets) with large kernels outperform similar models with standard small kernels on the task of nuclei segmentation in histopathology images. We observe that the large kernels mostly capture low-frequency information, which motivates the need for large kernels and their efficient compression via the Discrete Cosine Transform (DCT). Following this idea, we develop a U-Net model with wide and compressed DCT kernels that leads to similar performance and trends to the standard U-Net, with reduced complexity.
Open PhD position – Machine learning and multimedia knowledge extraction from biomedical data
We are looking for a talented PhD student in the MedGIFT research group.
The position involves working on machine learning and multimedia knowledge extraction from biomedical data.
The PhD will be part of the European Horizon research project HEREDITARY, made up of a consortium of 18 partners from Europe and the United States of America.
More information here.
Other PhD, post-doc and internship positions are also open, do not hesitate to contact us for more information.
Mara Graziani awarded prestigious Latsis prize at UNIGE
The Latsis University Prize aims to acknowledge outstanding work conducted by young researchers. Mara Graziani has been honored with this prestigious award in recognition of her remarkable contributions to the field of trustworthy AI. Her research has significantly enhanced the comprehensibility of deep learning models and their ability to generalize to unseen datasets.
Many of the current artificial intelligence (AI) algorithms operate as “black boxes,” meaning they lack transparency in explaining the reasoning behind their predictions. This lack of transparency poses significant challenges in the use and regulation of AI-based devices in high-stakes contexts.
During her Ph.D. at MedGIFT (HES-SO) and UNIGE, Mara developed several innovative methods that shed light on the inner workings of complex deep learning models. Additionally, she has made substantial progress in multi-task learning methods, guiding models to focus on essential features. This approach has proven to greatly enhance the models’ generalization capabilities and their resilience to domain shifts.
In addition to her remarkable research contributions, Mara was honored with the prize for her commitment to the AI community. In 2021, she initiated the “Introduction to Interpretable AI” expert network, aimed at fostering global discussions on deep learning interpretability. This initiative has reached hundreds of students worldwide and received support from various AI experts who have contributed through online seminars, lecture notes, and open-source code.
Link to the award ceremony here (38’45”)

Article on canine thoracic radiographs classification published in Scientific Reports
Our recent work on “An AI-based algorithm for the automatic evaluation of image quality in canine thoracic radiographs” has been published in Scientific Reports (Nature).
The aim of this study was to develop and test an artificial intelligence (AI)-based algorithm for detecting common technical errors in canine thoracic radiography. More specifically, the algorithm was designed to classify the images as correct or having one or more of the following errors: rotation, underexposure, overexposure, incorrect limb positioning, incorrect neck positioning, blurriness, cut-off, or the presence of foreign objects, or medical devices. The algorithm was able to correctly identify errors in thoracic radiographs with an overall accuracy of 81.5% in latero-lateral and 75.7% in sagittal images.

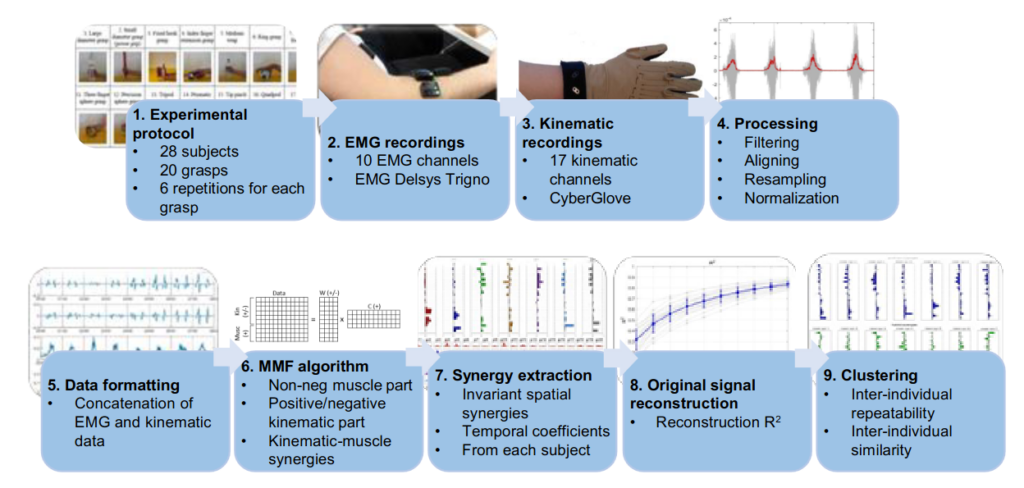
Article on Kinematic-Muscular Synergies in Hand Grasp Patterns Published in IEEE Access
Our recent work “Functional synergies applied to a publicly available dataset of hand grasps show evidence of kinematic-muscular synergistic control” has been published in IEEE access.
Hand grasp patterns are the results of complex kinematic-muscular coordination and
synergistic control might help reducing the dimensionality of the motor control space at the hand level.
Kinematic-muscular synergies combining muscle and kinematic hand grasp data have not been investigated before. This paper provides a novel analysis of kinematic-muscular synergies from kinematic-muscular data of 28 subjects, performing up to 20 hand grasps.
The results generalize the description of muscle and hand kinematics, better clarifying several limits of the field and fostering the development of applications in rehabilitation and assistive robotics.
Article published in Scientific Data (Nature)
Our latest paper published in Scientific Data (Nature), entitled “Multimodal video and IMU kinematic dataset on daily life activities using affordable devices”, M. Martínez-Zarzuela et al. , describes our publicly available dataset VIDIMU, available in Zenodo repository.
The objective of the VIDIMU dataset is to pave the way towards affordable patient gross motor tracking solutions for daily life activities recognition and kinematic analysis in out-of-the-lab environments.
The novelty of this dataset lies in: (i) the clinical relevance of the chosen movements, (ii) the combined utilization of affordable video and custom sensors, and (iii) the implementation of state-of-the-art tools for multimodal data processing of 3D body pose tracking and motion reconstruction in a musculoskeletal model from inertial data.


HECKTOR paper published in Medical Image Analysis
Our article “Automatic Head and Neck Tumor segmentation and outcome prediction relying on FDG-PET/CT images: Findings from the second edition of the HECKTOR challenge“, V. Andrearczyk et al., has been published in Medical Image Analysis.
We describe the second edition of the HECKTOR challenge, including the data, tasks and participation, and present various post-challenge analyses including ranking robustness, ensembles of algorithms, inter-center performance, and influence of tumor size on performance.

For participation in the latest challenge, go to this link.
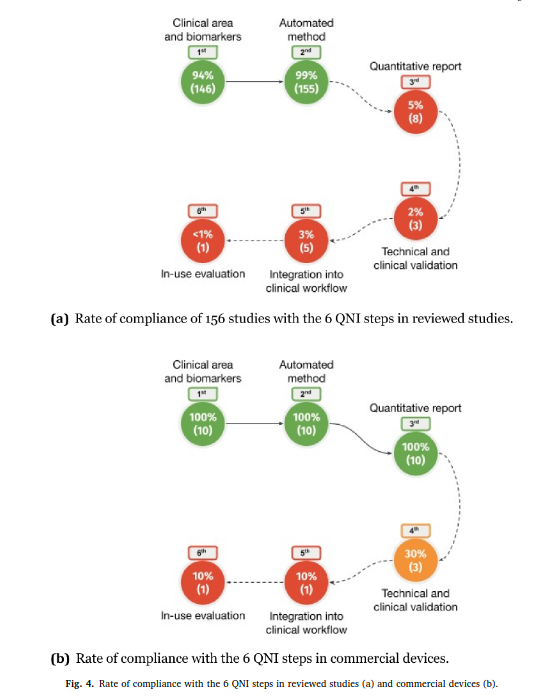
Review article published in NeuroImage: Clinical
Our review paper “Automated MS lesion detection and segmentation in clinical workflow: a systematic review” was published in NeuroImage: Clinical.
This work, conducted by Federico Spagnolo and colleagues, is the result of a great collaboration between HES-SO, CHUV and UniBasel. We investigate to what extent automatic tools in multiple sclerosis management fulfill the Quantitative Neuroradiology Initiative (QNI) framework necessary to integrate automated detection and segmentation into the clinical neuroradiology workflow.