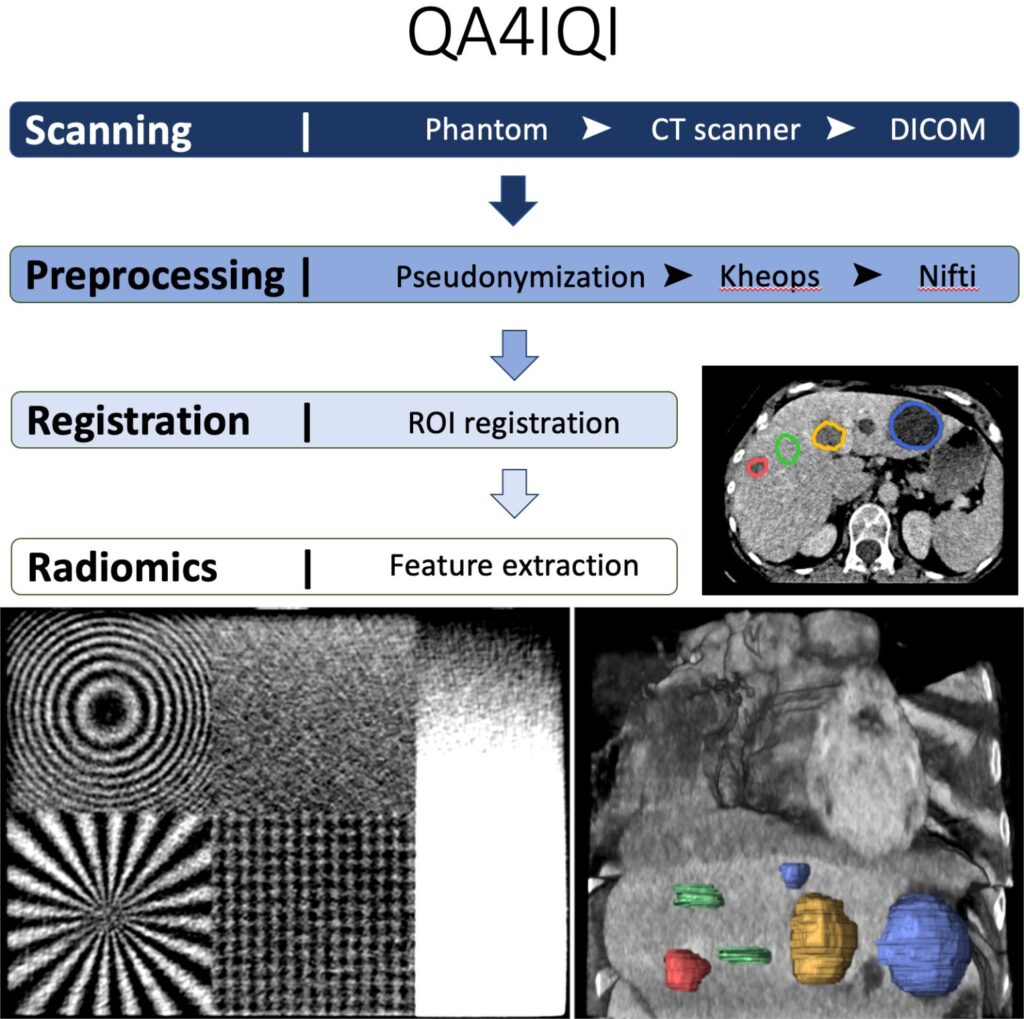
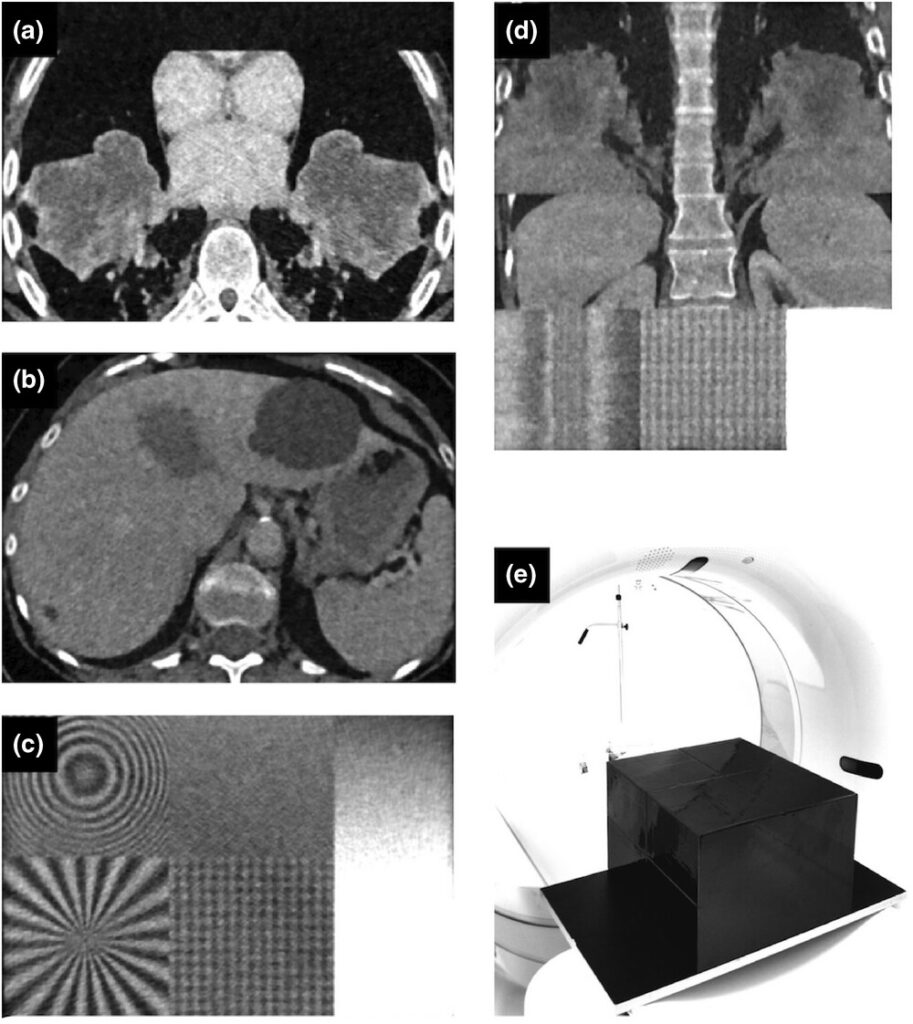
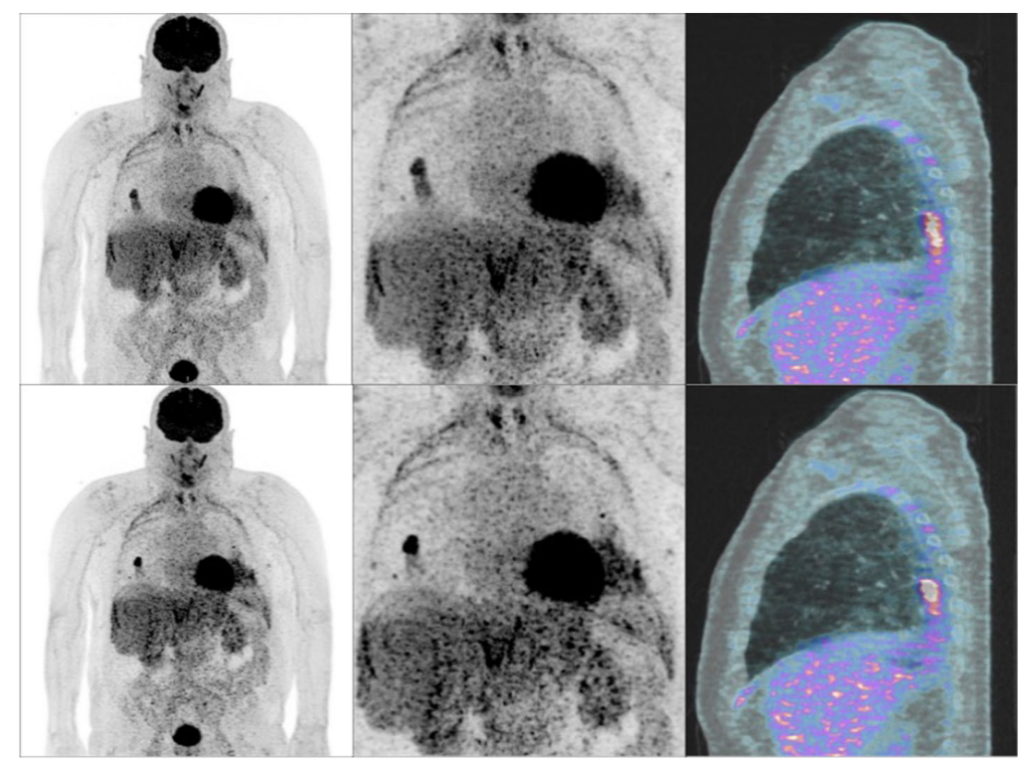
Our new dataset “Task-Based Anthropomorphic CT Phantom for Radiomics Stability and Discriminatory Power Analyses (CT-Phantom4Radiomics)” has been published on The Cancer Imaging Archive (TCIA). The aims of this dataset are to determine the stability of radiomics features against computed tomography (CT) parameter variations and to study their discriminative power concerning tissue classification using a 3D-printed CT phantom based on real patient data.
The dataset is available here: https://doi.org/10.7937/a1v1-rc66
The source code and instructions on how to run the full feature extraction pipeline on the TCIA dataset can be found here: https://github.com/QA4IQI/qa4iqi-extraction
More details on the QA4IQI project results: https://qa4iqi.github.io/